Showing the Power of Parquet and Ibis Using UK Census Data
By
K
Kae SuarezAuthor(s):
K
Kae SuarezPublished: July 20, 2023
8 min read
TL;DR Parquet is touted for its compression (storage) advantages and in this blog we show how converting your Comma Separated Value files (CSVs) to Parquet files will drastically increase your performance advantage at the trivial cost of a few lines of code.
Comma Separated Value files (CSVs) are ubiquitous and carry a strong legacy. Between human readability and parsers existing for almost any data tool, CSVs have been standard files for data. However, this ubiquity does not translate to usability or performance — or even flexibility. From disparate behaviors between parsers to the performance cost of parsing and converting strings to other data types, and a lack of nested types, CSV fails to meet enterprise demands. Yet, CSV ubiquity means many datasets are most easily found and hosted as CSV.
However, a powerful alternative is becoming commonplace. The Parquet format sacrifices human readability for usability, performance, and flexibility. With embedded metadata and a binary format, parsing is easy and efficient, and support for complex data is present. Adoption is rising quickly, and you can find Parquet support everywhere from DuckDB to pandas, allowing you to optimize performance for your connected data and processing systems. Further, where CSV is row-based, Parquet is columnar, enabling partial reads and columnar data in-memory, so you can augment your system to only perform exactly what you need, at the speed of columnar. Of course, these are just file formats — if you have the space, you can host both, so you never have to give up your existing structures.
Switching to Parquet is easy, especially with powerful Python tools like Ibis. Today, we’ll use Ibis and DuckDB to convert UK Census data from CSV to Parquet, enhancing usability and flexibility in a few easy steps.
UK Census
The UK Census publishes data in an online portal for building tables, but also packages all underlying bulk data in accessible CSVs. At 155 MB compressed in .zip files, and 590 MB uncompressed, what is interesting here is not size and bandwidth, but the shape of the data:
- 73 zip files
- Corresponding to 73 sets of related features
- Each containing 5 CSV files corresponding to geographical division methods
It can be intimidating at first but, for many use cases, only one or two sets are needed. However, as an application grows more complex and demands more diverse data, a user is left performing joins between tables read from these CSVs repeatedly — if you join all of them, you end up with 2189 columns. Having a single 2189 column-wide CSV is not an efficient use of storage resources, as this penalizes both storage capacity and read/write performance.
That’s where Parquet comes in. Offering partial file reads and, in this case, 5x better compression than the .zip files used here, Parquet files simplify the process of reading in everything you could need, without sacrificing the granularity of reading specific sets of columns.
First, lets download the UK census data. It is accessed via mostly sequential URLs. Unfortunately, there are gaps in the numbered zip files, which still have valid URLs — they simply download blank .zips. We will handle this when reading tables, and download and unzip the files naively for now.
To download:
bash
for i in {1..9}
do
wget https://www.nomisweb.co.uk/output/census/2021/census2021-ts00$i.zip
done
for i in {10..79}
do
wget https://www.nomisweb.co.uk/output/census/2021/census2021-ts0$i.zip
doneTo unzip:
bash
for i in {1..9}
do
unzip -d census2021-ts00$i census2021-ts00$i.zip
done
for i in {10..79}
do
unzip -d census2021-ts0$i census2021-ts0$i.zip
doneConverting to Parquet with Ibis
Now, we want to move from .zip files with five CSVs each to five Parquet files. While a join could also happen over the region types, keeping them separate is more in spirit with the original packaging where having different methods of dividing into regions is meaningful. So, we need to create five equivalent Parquet tables, each made up of data from every corresponding CSV.
For any given set of regions, we’ll use the regions therein as the keys for our join. However, not every CSV for a region set has all regions — some measurements simply are not taken in some areas. With this in mind, an inner join is inappropriate, as the resulting dataset would only include the keys present in all files. Outer joins solve this data dropping problem, but can and do cause column duplication.
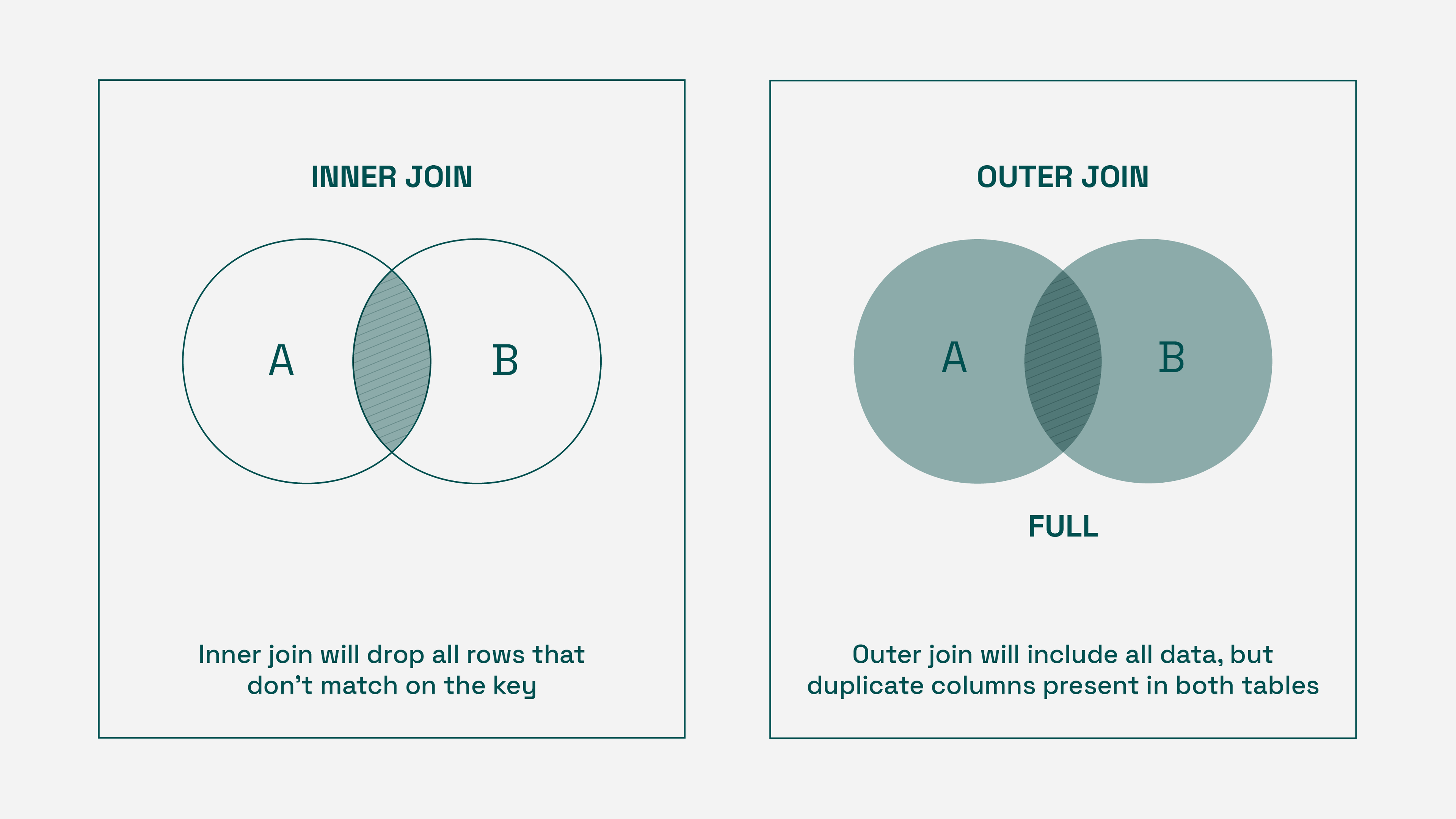
Thus, we’ll use an outer join, while dropping duplicate columns.
The flow will be as follows:
- For all region classifications:
- For all files:
- Read CSV
- For all tables:
- Join to master table
- Deduplicate columns
- Write out master table to Parquet
- For all files:
To implement, we’ll start with imports, and connect to DuckDB:
python
import ibis
from ibis import selectors as s
import pyarrow as pa
from pyarrow import parquet
import pyarrow.dataset as ds
import os
from ibis import _
con = ibis.duckdb.connect()Now, to define the function:
python
def to_parquet(region):
# We'll use an array to keep track of all our read files.
tables = []
for i in range(1, 80):
# Because all the URLs are valid but not all files, we check to see which
# files are real.
if os.path.exists(f"census2021-ts{i:03}/census2021-ts{i:03}-{region}.csv"):
tables.append(con.read_csv(f"census2021-ts{i:03}/census2021-ts{i:03}-{region}.csv", table_name=f"ts{i:03}").relabel("snake_case"))
# We start with the first table, then join into it repeatedly.
t = tables[0]
for t2 in tables[1:]:
# This expression joins on the date, geography, and geography code
# keys, then deduplicates the generated columns we don't need.
t = t.join(t2, predicates=("date", "geography","geography_code"), how="outer").mutate(
date = ibis.coalesce(_.date_x, _.date_y), geography = ibis.coalesce(_.geography_x, _.geography_y),
geography_code = ibis.coalesce(_.geography_code_x, _.geography_code_y))
t = t.drop("date_x", "geography_x", "geography_code_x", "date_y", "geography_y", "geography_code_y")
# Cache executes in database, because we do not need to manifest in Python.
out = t.cache()
# Output to Parquet for region type.
out.to_parquet(f"{region}_out.parquet")
Since there are only five region types, we can simply iterate over them with literals:
python
regions = ["ctry", "ltla", "msoa", "rgn", "utla"]
for region in regions:
to_parquet(region)This leaves us with Parquet files. What do we get out of that?
Benefits of Parquet for UK Census
One benefit of using Parquet is compression. Altogether the .zip files are around 160 MB on disk. Uncompressed, these files balloon to around 600 MB. When converted to Parquet, the resulting files are 26 MB in size. Between effective binary encoding and compression-by-default, Parquet can compress data very well, while avoiding the need to decompress by hand before using.
While this size of data easily fits in memory, the other benefit is in usability. The files are now very wide tables, and while this was the very situation the original structure we wanted to avoid, Parquet’s columnar nature, as opposed to row based format means the experience — and compute — are improved. With Parquet, you can select specific columns to be read, without the overhead of also reading other unnecessary columns along the way:
python
import pyarrow.parquet as pq
print(pq.read_table("ltla_out.parquet", columns=["geography_code", "passports_held:_total;_measures:_value"]))
OUT:
pyarrow.Table
geography_code: string
passports_held:_total;_measures:_value: int64
----
geography_code: [["E06000001","E06000002","E06000003","E08000022","E08000023",...,"E06000051","E07000106","E07000028","E07000061","E07000070"]]
passports_held:_total;_measures:_value: [[92339,143922,136529,208965,147774,...,323605,157430,110024,101686,181521]]
The code above did not drop extra columns. It is a feature of Parquet that, by storing columns instead of rows, and storing metadata as a standardized footer, a device can read only the metadata and requested data. To confirm, we provide an excerpt of the trace output for accesses on the LTLA file when read as a table versus by column, specifically where the data is read. The number on the right is the number of bytes read:
python
Table read:
[pid 5944] pread64(21, "\25\0\25\276)\25\350\31,\25\226\5\25\0\25\6\25\6\0\0\337\24(\3\0\0\0\226\5\1\262h"..., 2334049, 4) = 2334049
Column read:
[pid 5998] pread64(21, "\25\0\25\276)\25\320\31,\25\226\5\25\0\25\6\25\6\0\0\337\24(\3\0\0\0\226\5\1Oe"..., 1660, 1676) = 1660
When columns are read instead of rows, fewer bytes are ingested! A novelty and convenience at this scale, but essential for keeping data wrangling tractable when dealing with Big Data.
Conclusion
In the past, we have spoken about Parquet’s speed and compressibility. But it isn’t just the file format for speed — it’s for usability and flexibility as well, even on datasets with unique challenges, such as the UK Census dataset’s width. Starting with Parquet is not difficult either: simply load your data into your tool of choice, then write it out to Parquet format. You can even tune your schema to maximize gains like we did with the U.S. Census PUMS dataset.
Using Parquet also doesn’t force you to give anything up. Many of the tools you love, like pandas, Polars, or Ibis, ship with Parquet and CSV compatibility, so you can just swap your ingest phase and enjoy the benefits of the columnar file format.
Many organizations, government agencies, and enterprises have servers full of CSV data that are impacting system performance. Voltron Data helps organizations design and build data systems by augmenting what they have using open source standards like Parquet and tools like Ibis. If you’re interested in learning more about our products and approach, click here.
Photo by Lachlan Gowen
Previous Article
Keep up with us



