Ibis v3.2.0 Brings More Ways to Tackle Tabular Data
By
M
Marlene MhangamiAuthor(s):
M
Marlene MhangamiPublished: September 29, 2022
9 min read
The Ibis v3.2.0 release brings a new wave of tools to help developers work with tabular data. This post highlights the top three features that come with this release and clarifies how — and why — to use them. A full list of changes in 3.2.0 can be found in the project’s release notes.
New Features
Ibis 3.2.0 introduces many new features and improvements that both align to the roadmap and respond to the community’s requests. These are our three standout feature picks:
- New Memtable Feature to Speed up Pandas with DuckDB
- DuckDB is Now the Default Ibis Backend
- New Struct Unpacking for Working with Nested Data
Refer to the sections below for more detail on each feature.
New Memtable Feature to Speed up Pandas with DuckDB
Memtable is a feature that gives Ibis users the ability to interact with Pandas dataframes as Ibis expressions in a convenient way. Memtable takes in a dataframe and creates an Ibis expression that stores it and can have SQL queries run on it. With larger datasets (100 million rows and above) running queries on dataframes this way becomes significantly more performant than using Pandas for the same operations. Currently, DuckDB is the default backend, with ClickHouse being natively supported as well. Let’s see some examples of what this looks like with code.
We start off by reading data from the New York City Taxi Dataset for the month of February and storing it in a Pandas dataframe.
python
import ibis
import pandas as pd
df= pd.read_csv(‘yellow_tripdata_2009-02.csv’)
df.head()Run the code yourself in the accompanying notebook for this blog found here.
Next, let’s pass the dataframe to memtable so it’s accessible as an Ibis expression. We’ll also set interactive mode to false so we can see more details about the ibis expression. Take a look at the code below along with a picture of output after the dataframe is passed to memtable.
python
ibis.options.interactive = False
ibis.memtable(df)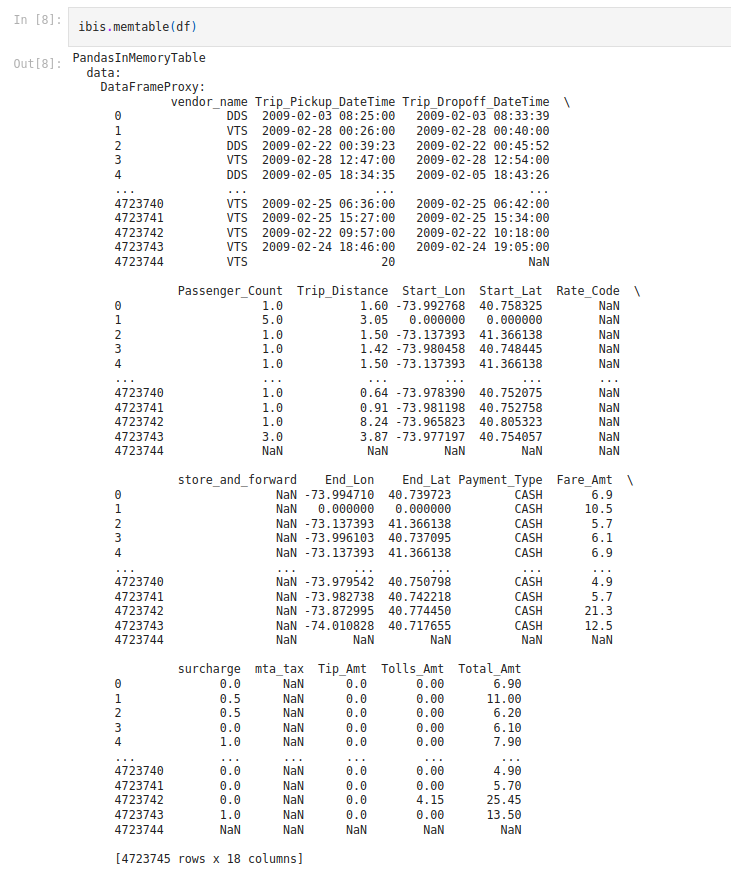
Figure 1: In-memory table produced after calling memtable.
What we see returned is an ‘InMemoryTable’ that can conveniently have SQL queries run over it. Let’s look at some example code of different operations being called on the table.
python
t = ibis.memtable(df)
ibis.options.interactive = False
t.columns
t.vendor_name.count()We first save the table to a variable t and set interactive mode to False so that we don’t get as much detail or need to call execute. Next, we call t.columns to get a better understanding of the information available. Since the dataframe is now an Ibis expression we can find the number of vendors by running t.vendor_name.count(). We find that there are 4,723,745 taxi vendors!
It might be helpful to understand how this is different from calling the same operation in Pandas. In order to visualize we can use ibis.show_sql to see what’s happening under the hood. Let’s calculate the sum of the surcharge column and take a look at the SQL code.

Figure 2: showing the SQL code DuckDB runs when the operation for calculating the surcharge sum is run in Ibis
We see that an SQL select statement is being called on _ibis_memtable1. The table being queried, _ibis_memtable1, is actually a temporary view that Ibis creates using native facilities in DuckDB to represent the in-memory table. DuckDB is able to fully execute SQL queries against our in-memory dataframe!
Performance
As mentioned earlier, one of the advantages of using memtable is the speed up you get with larger datasets compared to Pandas. Let’s make a quick comparison of the two using some generated data. We use the following code to generate a Pandas dataframe containing 100 million rows.
python
df_large = pd.DataFrame({
"a": np.random.randn(int(1e8)),
"b": ['a','b'] * int(1e8 // 2)
}).astype({"b": "string"})We are mainly concerned about the number of rows, as compared to the table content, to clearly see performance differences.
Next, we’ll measure how long it takes to do a value count on column ‘b’ with Pandas. To do this we will call the value_counts method on it. I’ve taken a screenshot of my results but you can test it yourself in the notebook.
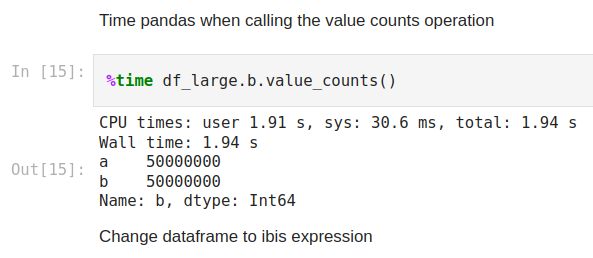
Figure 3
Here we can see that the Wall time for the operation to be complete is 1.94s. Now let’s compare this to Ibis where we use memtable to read the dataframe into memory and query it with DuckDB. We’ll first convert the Pandas dataframe to an in-memory table using memtable and then run the same operation and time assessment as shown below.
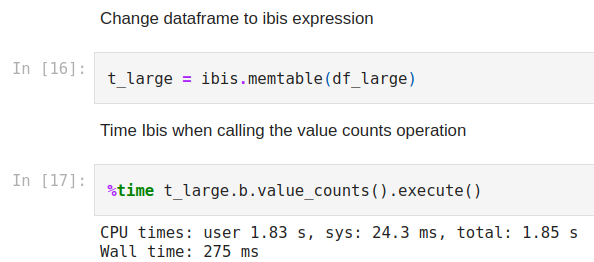
Figure 4
As we can see above this only took 275ms. This means using Ibis to carry out this operation was almost 8X faster than Pandas!
Users can also use memtables to join Pandas dataframes to Ibis expressions. This allows for quick and easy mixing of local data and backend data: rather than uploading ad-hoc or random files to your backend, you can simply read them in as a dataframe and join.
As we can see, using memtable provides a range of benefits to Ibis users. For more information on how to use this feature check out Ibis maintainer Phillip Cloud sharing more here.
DuckDB is the Default Ibis Backend
For features like memtable and others where users have the option to not explicitly choose a backend, Ibis now defaults to DuckDB. The DuckDB backend was chosen primarily because it supports the widest variety of SQL operations of the Ibis backend group. A table comparing the different Ibis backends based on which they support can be viewed [here](https://ibis-project.org/backends/support_matrix/ {:target=”_blank”}.
DuckDB also happens to be fast, as we saw in the example earlier with memtable. It can operate on PyArrow tables, record batches, Parquet files, etc. It also has a feature that can register an in memory dataframe and execute SQL queries on that memory natively. This makes it ideal for a case like memtable in Ibis. When an Ibis user calls ibis.memtable on a dataframe an in-memory DuckDB connection is created. This connection is not tied to any DuckDB database, so the connection goes away when your Python process exits. This ephemeral nature is great for things that are operating in-memory since having a new database formed or a large random file generated would not be ideal! When an expression is called on the in-memory table, Ibis detects whether or not that connection can be executed by the in-memory connection. If it can, the corresponding SQL query will run on the table and the results are returned.
It’s important to note that if a user already selected a backend before calling something like memtable, nothing will change and that backend will be used. DuckDB will only be used when the expression does not have an existing backend and it can be executed. It is also possible to change the default backend to something other than DuckDB using the following line of code ibis.options.default_backend. An example of setting the default backend to Pandas is ibis.options.default_backend = “pandas.
For more information on the choice of DuckDB as the default watch lead Ibis maintainer, Phillip Cloud, and I discuss it briefly on our YouTube Channel.
New Struct Unpacking for Working with Nested Data
Finally, the last major feature added in 3.2 is struct unpacking. This method is useful when analyzing a data set that has a lot of nested structs or arrays of structs because it allows you to chain unpack method calls and avoid repeating column names and tables in an expression.
Let’s take a quick look at a code example to see what this looks like practically. Below is the code needed to create a table from a dictionary with two keys ‘a’ and ‘d’. Column ‘a’ is made up of nested data, and more specifically contains a struct that in itself has two fields ‘b’ and ‘c’. We can then run unpack on column ‘a’ and the resulting table can be seen and compared to the original one in the image below.
python
t = ibis.table(dict(a="struct", d="string"), name="t")
t.unpack(‘a’)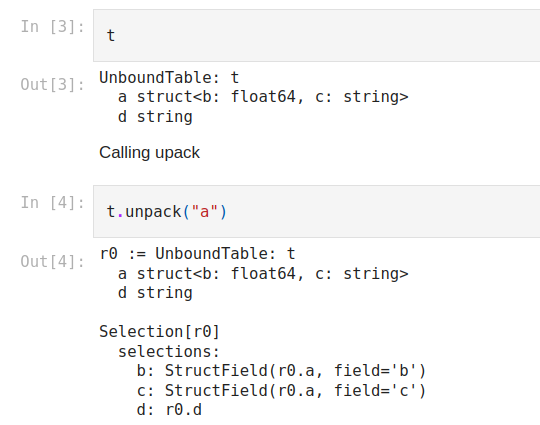
Figure 5
When we call the unpack method on column ‘a’, the fields of the struct from ‘a’ become individual columns. The information shown in each of the new columns lets us know that they originated from the table r0 (which is table t) and the first column, r0.a, along with their field names. Being able to unpack structs in this way makes your data easier to work with and saves time.
As mentioned earlier, when working with larger data sets with more complex nesting, users can chain struct method calls to unpack several layers of structs. To learn more about what this looks like with real-world data you can watch a live demo of unpack.
To access the code and data from this post check out the notebooks.
Conclusion
In summary, our favorite features from this release include the ability to use and run queries on dataframes in Ibis, DuckDB, a feature-rich and performant backend becoming the default backend, and ibis.unpack providing a more convenient way to work with highly nested data.
Ibis is a community-led, open source project. If you’d like to contribute to the project check out the contribution guide. If you run into a problem and would like to submit an issue, you can do so through Ibis’ Github repository. Finally, Ibis relies on community support to grow and become successful! You can help promote Ibis by following and sharing the project on Twitter, starring the repo, contributing to the code, or subscribing on YouTube. Ibis continues to improve with every release. Keep an eye on the blog for updates on the next one!
For even more resources, learn how a Voltron Data Enterprise Support subscription can help accelerate your success with Ibis.
Photo by Ruslan Alekso
Previous Article
Keep up with us



